Scripts & Experiments
Table of Contents
Calling scripts from the command line
Working on compute-clusters often required scheduling "jobs" from the command-line. To run a Julia script in the file my_script.jl, run the following command:
$ julia my_script.jl arg1 arg2...Inside your script, the additional command-line arguments arg1 and arg2 can be used through the global constant ARGS. If my_script.jl contains the code
# Content of my_script.jl
for a in ARGS
println(a)
endCalling it with arguments foo, bar from the command-line will print:
$ julia my_script.jl foo bar
foo
barCommand-line switches
Julia provides several command-line switches. For example, for parallel computing, --threads can be used to specify the number of CPU threads and --procs for the number of worker processes.
The following command will run my_script.jl with 8 threads:
$ julia --threads 8 -- my_script.jl arg1 arg2Parallel computing
In this lecture, we only covered GPU parallelization (Lecture 7 on Deep Learning).
Refer to the Julia documentation on parallel computing for more information on multi-threading and distributed computing.
External packages
Handling arguments in ARGS can be tedious. Comonicon.jl is a package to build simple command-line interfaces for Julia programs by using a macro @main. Among other features, it supports
positional arguments
optional arguments with defaults
boolean flags
help pages generated from docstrings
Take a look at the documentation.
Experiments with DrWatson.jl
DrWatson.jl describes itself as "scientific project assistant software". It serves two purposes:
It sets up a project structure that is specialized for scientific experiments, similar to PkgTemplates.
It introduces several useful helper functions. Among these are boiler-plate functions for file loading and saving.
The following two sections are directly taken from the DrWatson documentation, which I recommend reading
File structure
To initialize a DrWatson project, run:
julia> using DrWatson
julia> initialize_project("MyScientificProject"; authors="Adrian Hill", force=true)The default setup will initialize a file structure that looks as follows:
│projectdir <- Project's main folder. It is initialized as a Git
│ repository with a reasonable .gitignore file.
│
├── _research <- WIP scripts, code, notes, comments,
│ | to-dos and anything in an alpha state.
│ └── tmp <- Temporary data folder.
│
├── data <- **Immutable and add-only!**
│ ├── sims <- Data resulting directly from simulations.
│ ├── exp_pro <- Data from processing experiments.
│ └── exp_raw <- Raw experimental data.
│
├── plots <- Self-explanatory.
├── notebooks <- Jupyter, Weave or any other mixed media notebooks.
│
├── papers <- Scientific papers resulting from the project.
│
├── scripts <- Various scripts, e.g. simulations, plotting, analysis,
│ │ The scripts use the `src` folder for their base code.
│ └── intro.jl <- Simple file that uses DrWatson and uses its greeting.
│
├── src <- Source code for use in this project. Contains functions,
│ structures and modules that are used throughout
│ the project and in multiple scripts.
│
├── test <- Folder containing tests for `src`.
│ └── runtests.jl <- Main test file, also run via continuous integration.
│
├── README.md <- Optional top-level README for anyone using this project.
├── .gitignore <- by default ignores _research, data, plots, videos,
│ notebooks and latex-compilation related files.
│
├── Manifest.toml <- Contains full list of exact package versions used currently.
└── Project.toml <- Main project file, allows activation and installation.
Includes DrWatson by default.Workflow
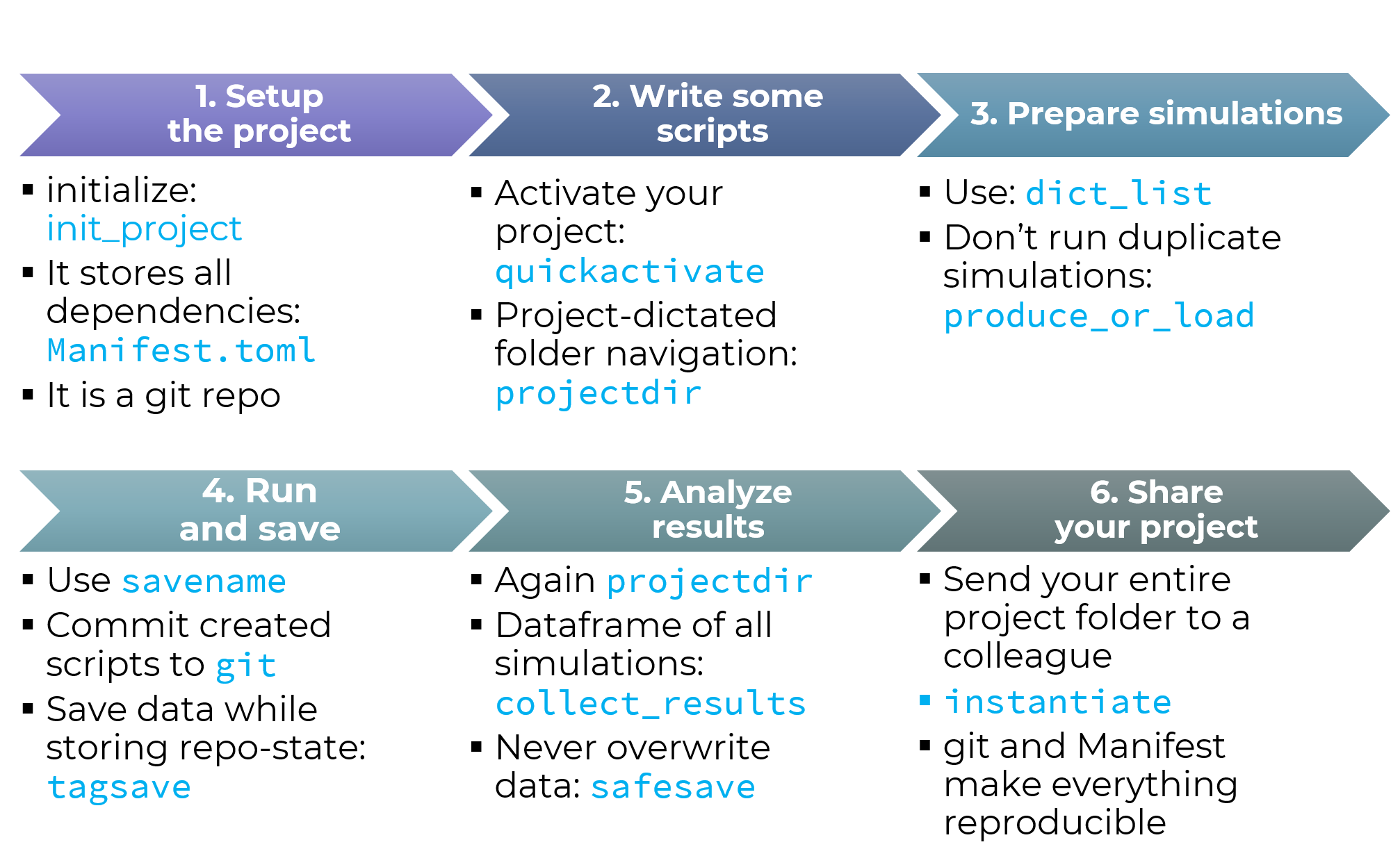
The DrWatson workflow is best summarized in the following picture from the documentation:
Website, code and notebooks are under MIT License © Adrian Hill.
Built with Franklin.jl, Pluto.jl and the Julia programming language.